UncommonBonds
SCIENCE
Our Approach
We use a covalent-first chemoproteomics approach to discover and develop new therapeutics that bind challenging-to-drug disease-causing protein targets. Through this approach we have uncovered hundreds of druggable pockets on protein targets with validated biology and links to disease as well as compounds that interact with these pockets in a highly selective manner. We have successfully progressed multiple candidates from initial screening to clinical trials and have a deep and rich pipeline of programs poised for future clinical entry.
VIVIDION’S covalent-first approach
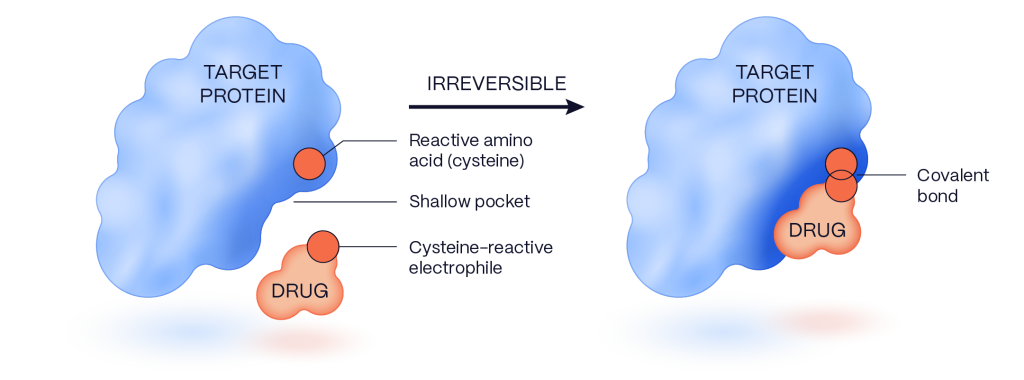
Our work uses covalent fragment libraries to identify protein sites amenable to forming irreversible bonds that are potent and selective. This covalent-first approach capitalizes on shallow pockets and requires only a small contact surface, which allows us to bind almost any disease-relevant target, including those that have eluded traditional drug discovery approaches, like transcription factors and ubiquitin ligases.
traditional drug discovery APPROACH
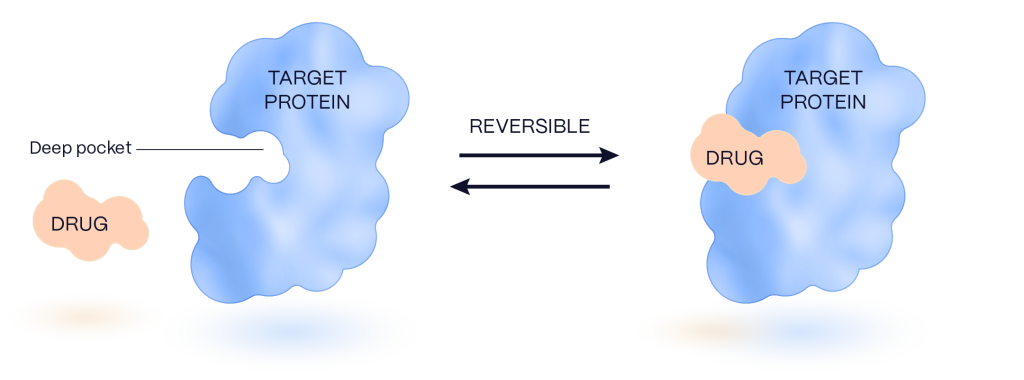
Much of the traditional small molecule discovery approach identifies protein sites that are bound potently, but not selectively or permanently. This approach is limited to proteins that interact with small molecule metabolites via deep pockets and form a large contact surface between the small molecule and the protein, which limits the number of druggable, potential disease-causing protein targets.
PLATFORM
Our innovation engine
Powering our approach is our proprietary library of covalent small molecules, an industrialized chemoproteomics screening platform, and an integrated data portal that together evaluate small molecule reactivity across the entire human proteome in an unbiased manner. Continuous, iterative library expansion allows for pipeline growth and a durable competitive advantage, with the potential to yield first-in-class and best-in-class medicines.
1. Library
Our custom-made library of over 50,000 covalent small molecules has been iteratively built to exploit novel electrophiles and innovative chemistry. These fragments are designed to be unreactive until they encounter a complementary protein surface on the disease target with a focus on reactive cysteines.
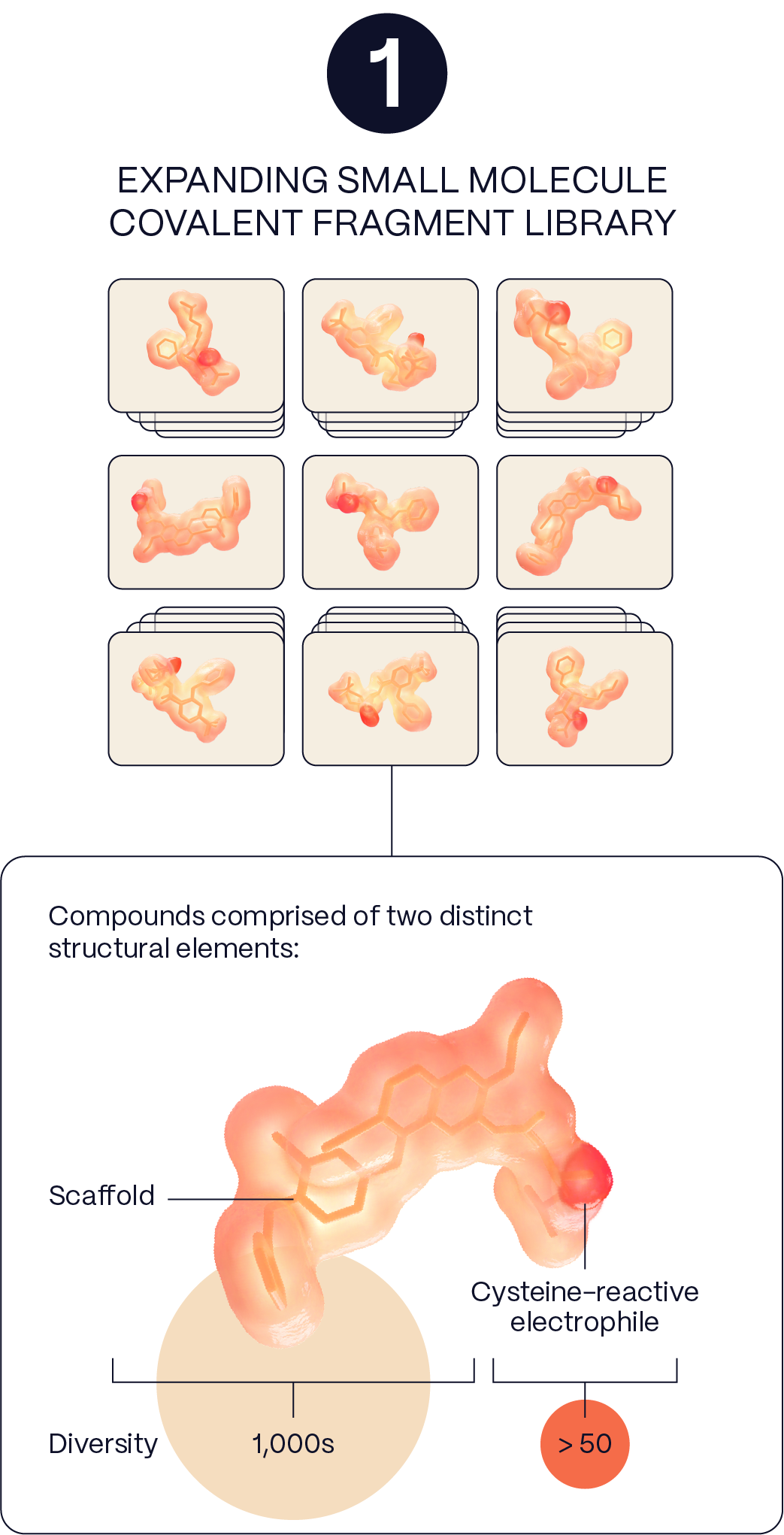
2. Screening
& Iterative Learning
We use high-resolution, high-throughput mass spectrometry to probe the surfaces of thousands of proteins in parallel in their native cellular state. This approach can reveal druggable pockets that may otherwise be hidden and provides real-time assessments of proteome-wide selectivity as hits advance.
3. Integrated Data Portal
Our data portal overlays the approximately 250,000 new data points generated per day by our chemoproteomics platform with the latest public resources for human genetics and structural biology. Integrating these data streams allows us to generate unique insights into accessible pockets and focus our discovery efforts on previously undruggable targets.
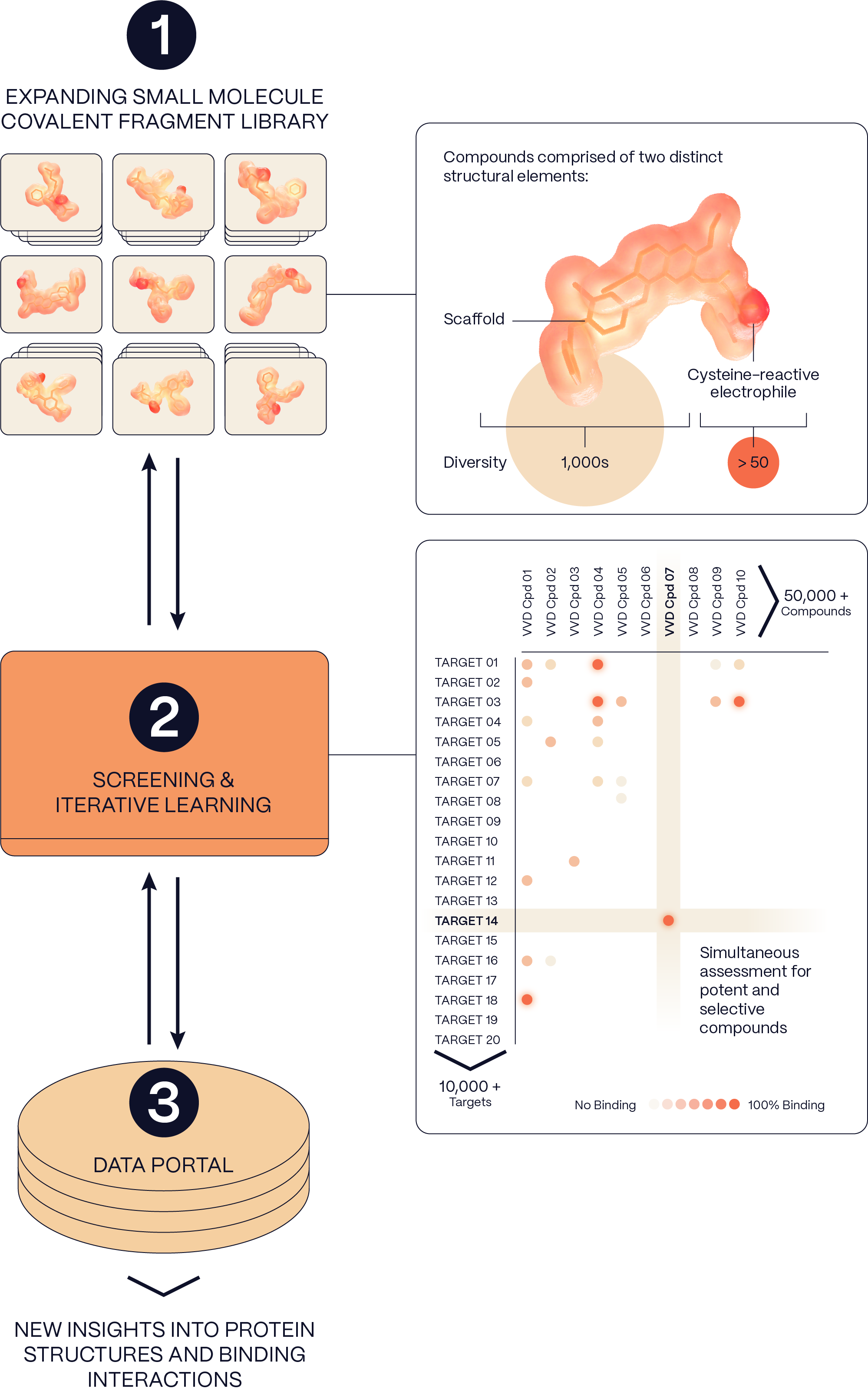
1. Library
Our custom-made library of over 50,000 covalent small molecules has been iteratively built to exploit novel electrophiles and innovative chemistry. These fragments are designed to be unreactive until they encounter a complementary protein surface on the disease target with a focus on reactive cysteines.
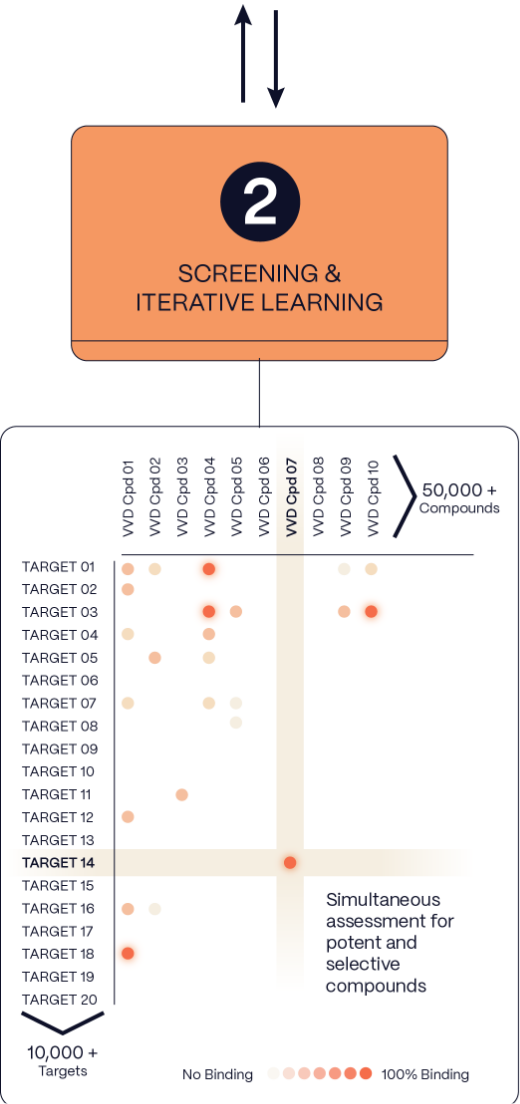
2. Screening
& Iterative Learning
We use high-resolution, high-throughput mass spectrometry to probe the surfaces of thousands of proteins in parallel in their native cellular state. This approach can reveal druggable pockets that may otherwise be hidden, and provides real-time assessments of proteome-wide selectivity as hits advance.
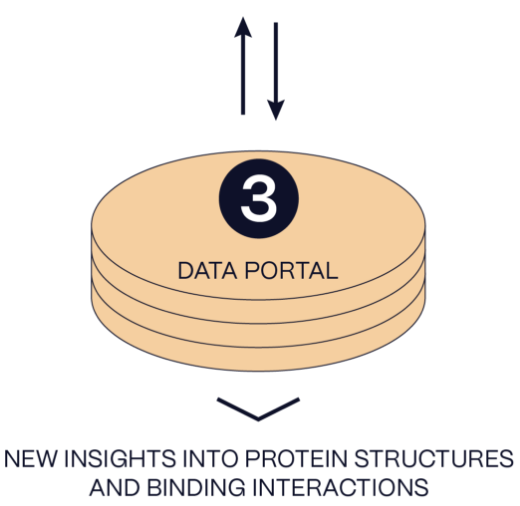
3. Integrated Data Portal
Our data portal overlays the approximately 250,000 new data points generated per day by our chemoproteomic platform with the latest public resources for human genetics and structural biology. Integrating these data streams allows us to generate unique insights into accessible pockets and focus our discovery efforts on previously undruggable targets.
This approach generates novel opportunities and greatly accelerates the drug development process by:
- quickly assessing many targets
- generating rich high-quality data on millions of interactions
- measuring potency and selectivity simultaneously
- emphasizing drug-like molecules
- identifying new allosteric- or cryptic-binding pockets, mechanisms
- uncovering novel mechanisms of action and generating new biological hypotheses
With our chemoproteomics approach, the commitment to a target builds as we screen and generate data, and we are able to screen an almost unlimited array of targets across any and all drug classes.
Broad therapeutic opportunity
To develop unique medicines that can impact disease biology in novel and therapeutically beneficial ways we are:
- uncovering previously unknown mechanisms of action and developing new biological hypotheses, even on disease targets with previously known biology, that guide our drug development
- finding molecules that irreversibly bind targets that were previously considered undruggable
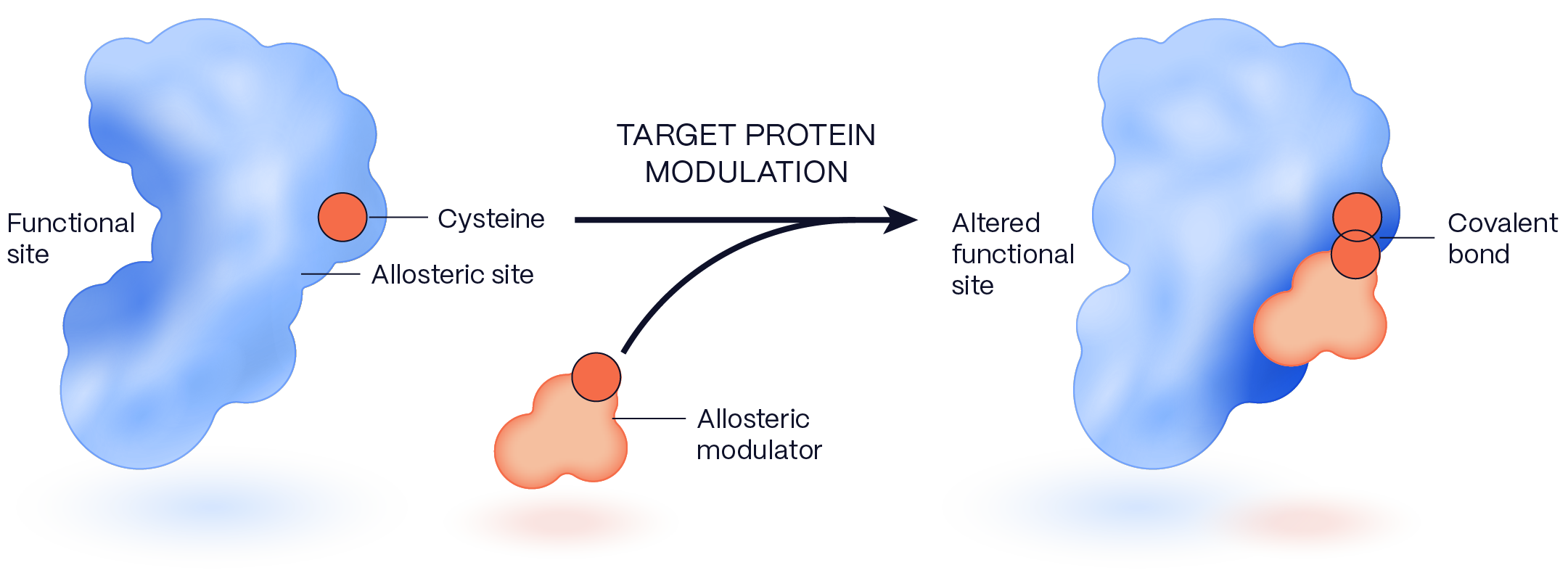
- identifying multiple ways to modulate these targets (inhibit or activate) by impacting protein function through allosteric sites, protein-protein interaction interfaces, or through induced proximity approaches including targeted protein degradation
ALLOSTERIC MODULATORS CAN MODIFY TARGET PROTEINS IN DIVERSE WAYS